Publications
Please refer to Google Scholar for more info.
2025
- Deep Biomechanically-Guided Interpolation for Keypoint-Based Brain Shift RegistrationTiago Assis*, Ines P Machado, Benjamin Zwick, Nuno Garcia, Reuben DorentWorkshop COLlaborative Intelligence and Autonomy in Image-guided Surgery (COLAS) at MICCAI 2025, 2025
Accurate compensation of brain shift is critical for maintaining the reliability of neuronavigation during neurosurgery. While keypoint-based registration methods offer robustness to large deformations and topological changes, they typically rely on simple geometric interpolators that ignore tissue biomechanics to create dense displacement fields. In this work, we propose a novel deep learning framework that estimates dense, physically plausible brain deformations from sparse matched keypoints. We first generate a large dataset of synthetic brain deformations using biomechanical simulations. Then, a residual 3D U-Net is trained to refine standard interpolation estimates into biomechanically guided deformations. Experiments on a large set of simulated displacement fields demonstrate that our method significantly outperforms classical interpolators, reducing by half the mean square error while introducing negligible computational overhead at inference time. Code available at: \hrefhttps://github.com/tiago-assis/Deep-Biomechanical-Interpolatorhttps://github.com/tiago-assis/Deep-Biomechanical-Interpolator.
@article{assis2025deep, title = {Deep Biomechanically-Guided Interpolation for Keypoint-Based Brain Shift Registration}, author = {Assis, Tiago and Machado, Ines P and Zwick, Benjamin and Garcia, Nuno and Dorent, Reuben}, journal = {Workshop COLlaborative Intelligence and Autonomy in Image-guided Surgery (COLAS) at MICCAI 2025}, year = {2025}, max_author_limit = {10} } - Unsupervised anomaly detection using Bayesian flow networks: application to brain FDG PET in the context of Alzheimer’s diseaseHugues Roy*, Reuben Dorent, and Ninon BurgosDeep Generative Models Workshop at MICCAI 2025, 2025
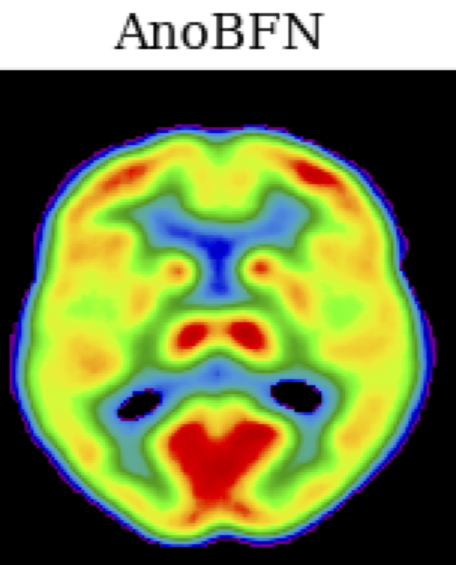
Unsupervised anomaly detection (UAD) plays a crucial role in neuroimaging for identifying deviations from healthy subject data and thus facilitating the diagnosis of neurological disorders. In this work, we focus on Bayesian flow networks (BFNs), a novel class of generative models, which have not yet been applied to medical imaging or anomaly detection. BFNs combine the strength of diffusion frameworks and Bayesian inference. We introduce AnoBFN, an extension of BFNs for UAD, designed to: i) perform conditional image generation under high levels of spatially correlated noise, and ii) preserve subject specificity by incorporating a recursive feedback from the input image throughout the generative process. We evaluate AnoBFN on the challenging task of Alzheimer’s disease-related anomaly detection in FDG PET images. Our approach outperforms other state-of-the-art methods based on VAEs (beta-VAE), GANs (f-AnoGAN), and diffusion models (AnoDDPM), demonstrating its effectiveness at detecting anomalies while reducing false positive rates.
@article{roy2025unsupervised, title = {Unsupervised anomaly detection using Bayesian flow networks: application to brain FDG PET in the context of Alzheimer's disease}, author = {Roy, Hugues and Dorent, Reuben and Burgos, Ninon}, journal = {Deep Generative Models Workshop at MICCAI 2025}, year = {2025} } - Unified Cross-Modal Medical Image Synthesis with Hierarchical Mixture of Product-of-ExpertsReuben Dorent*, Nazim Haouchine, Alexandra Golby, and 3 more authorsIEEE Transactions on Pattern Analysis & Machine Intelligence, 2025
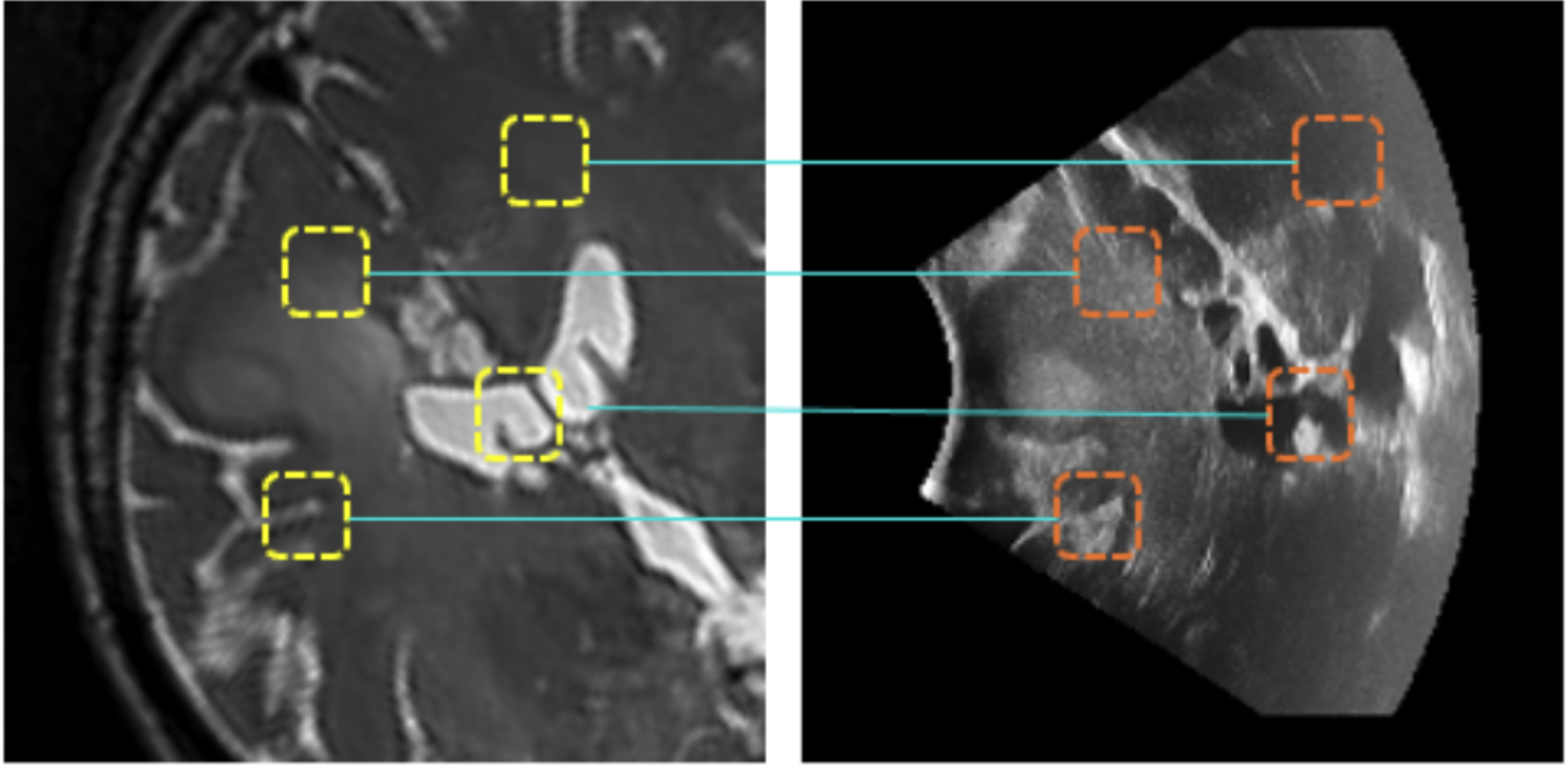
@article{dorent2025unified, title = {Unified Cross-Modal Medical Image Synthesis with Hierarchical Mixture of Product-of-Experts}, author = {Dorent, Reuben and Haouchine, Nazim and Golby, Alexandra and Frisken, Sarah and Kapur, Tina and Wells, William}, journal = {IEEE Transactions on Pattern Analysis \& Machine Intelligence}, year = {2025}, publisher = {IEEE Computer Society} } - A 3D Cross-modal Keypoint Descriptor for MR-US Matching and RegistrationDaniil Morozov*, Reuben Dorent, and Nazim HaouchinearXiv preprint arXiv:2507.18551, 2025
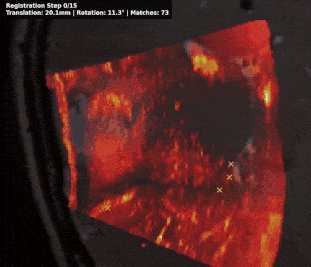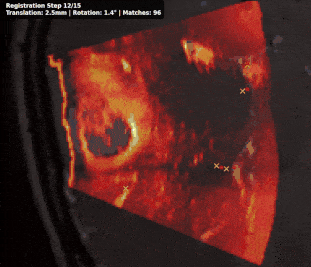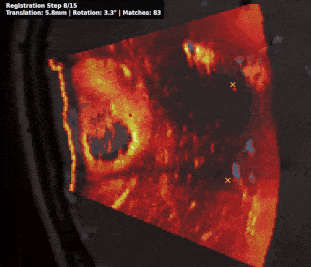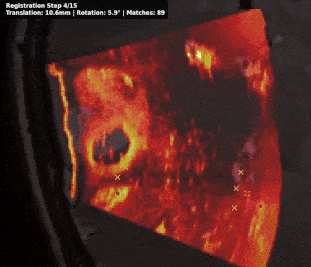
Intraoperative registration of real-time ultrasound (iUS) to preoperative Magnetic Resonance Imaging (MRI) remains an unsolved problem due to severe modality-specific differences in appearance, resolution, and field-of-view. To address this, we propose a novel 3D cross-modal keypoint descriptor for MRI-iUS matching and registration. Our approach employs a patient-specific matching-by-synthesis approach, generating synthetic iUS volumes from preoperative MRI. This enables supervised contrastive training to learn a shared descriptor space. A probabilistic keypoint detection strategy is then employed to identify anatomically salient and modality-consistent locations. During training, a curriculum-based triplet loss with dynamic hard negative mining is used to learn descriptors that are i) robust to iUS artifacts such as speckle noise and limited coverage, and ii) rotation-invariant . At inference, the method detects keypoints in MR and real iUS images and identifies sparse matches, which are then used to perform rigid registration. Our approach is evaluated using 3D MRI-iUS pairs from the ReMIND dataset. Experiments show that our approach outperforms state-of-the-art keypoint matching methods across 11 patients, with an average precision of . For image registration, our method achieves a competitive mean Target Registration Error of 2.39 mm on the ReMIND2Reg benchmark. Compared to existing iUS-MR registration approach, our framework is interpretable, requires no manual initialization, and shows robustness to iUS field-of-view variation. Code is available at https://github.com/morozovdd/CrossKEY.
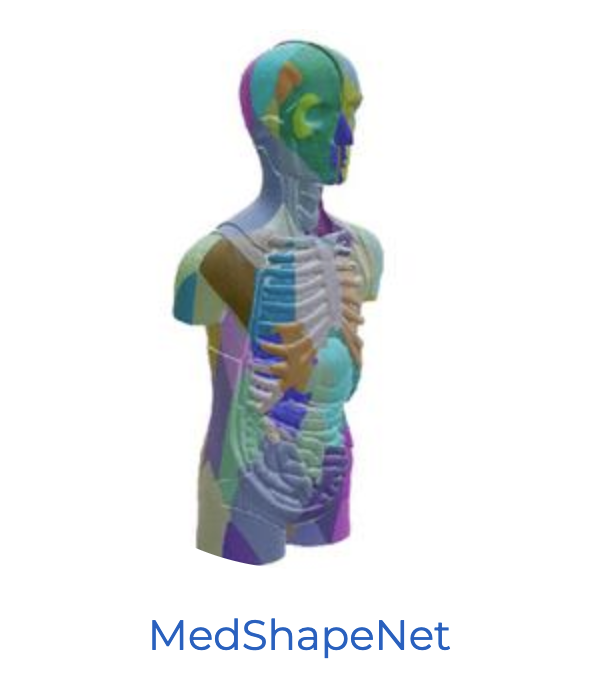
@article{morozov20253d, title = {A 3D Cross-modal Keypoint Descriptor for MR-US Matching and Registration}, author = {Morozov, Daniil and Dorent, Reuben and Haouchine, Nazim}, journal = {arXiv preprint arXiv:2507.18551}, year = {2025} } - Medshapenet–a large-scale dataset of 3d medical shapes for computer visionJianning Li*, Zongwei Zhou, Jiancheng Yang, and 8 more authorsBiomedical Engineering/Biomedizinische Technik, 2025
@article{li2025medshapenet, title = {Medshapenet--a large-scale dataset of 3d medical shapes for computer vision}, author = {Li, Jianning and Zhou, Zongwei and Yang, Jiancheng and Pepe, Antonio and Gsaxner, Christina and Luijten, Gijs and Qu, Chongyu and Zhang, Tiezheng and Chen, Xiaoxi and Li, Wenxuan and others}, journal = {Biomedical Engineering/Biomedizinische Technik}, volume = {70}, number = {1}, pages = {71--90}, year = {2025}, publisher = {de Gruyter}, } - crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023Navodini Wijethilake*, Reuben Dorent*, Marina Ivory, and 8 more authorsarXiv preprint arXiv:2506.12006, 2025
The cross-Modality Domain Adaptation (crossMoDA) challenge series, initiated in 2021 in conjunction with the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), focuses on unsupervised cross-modality segmentation, learning from contrast-enhanced T1 (ceT1) and transferring to T2 MRI. The task is an extreme example of domain shift chosen to serve as a meaningful and illustrative benchmark. From a clinical application perspective, it aims to automate Vestibular Schwannoma (VS) and cochlea segmentation on T2 scans for more cost-effective VS management. Over time, the challenge objectives have evolved to enhance its clinical relevance. The challenge evolved from using single-institutional data and basic segmentation in 2021 to incorporating multi-institutional data and Koos grading in 2022, and by 2023, it included heterogeneous routine data and sub-segmentation of intra- and extra-meatal tumour components. In this work, we report the findings of the 2022 and 2023 editions and perform a retrospective analysis of the challenge progression over the years. The observations from the successive challenge contributions indicate that the number of outliers decreases with an expanding dataset. This is notable since the diversity of scanning protocols of the datasets concurrently increased. The winning approach of the 2023 edition reduced the number of outliers on the 2021 and 2022 testing data, demonstrating how increased data heterogeneity can enhance segmentation performance even on homogeneous data. However, the cochlea Dice score declined in 2023, likely due to the added complexity from tumour sub-annotations affecting overall segmentation performance. While progress is still needed for clinically acceptable VS segmentation, the plateauing performance suggests that a more challenging cross-modal task may better serve future benchmarking.
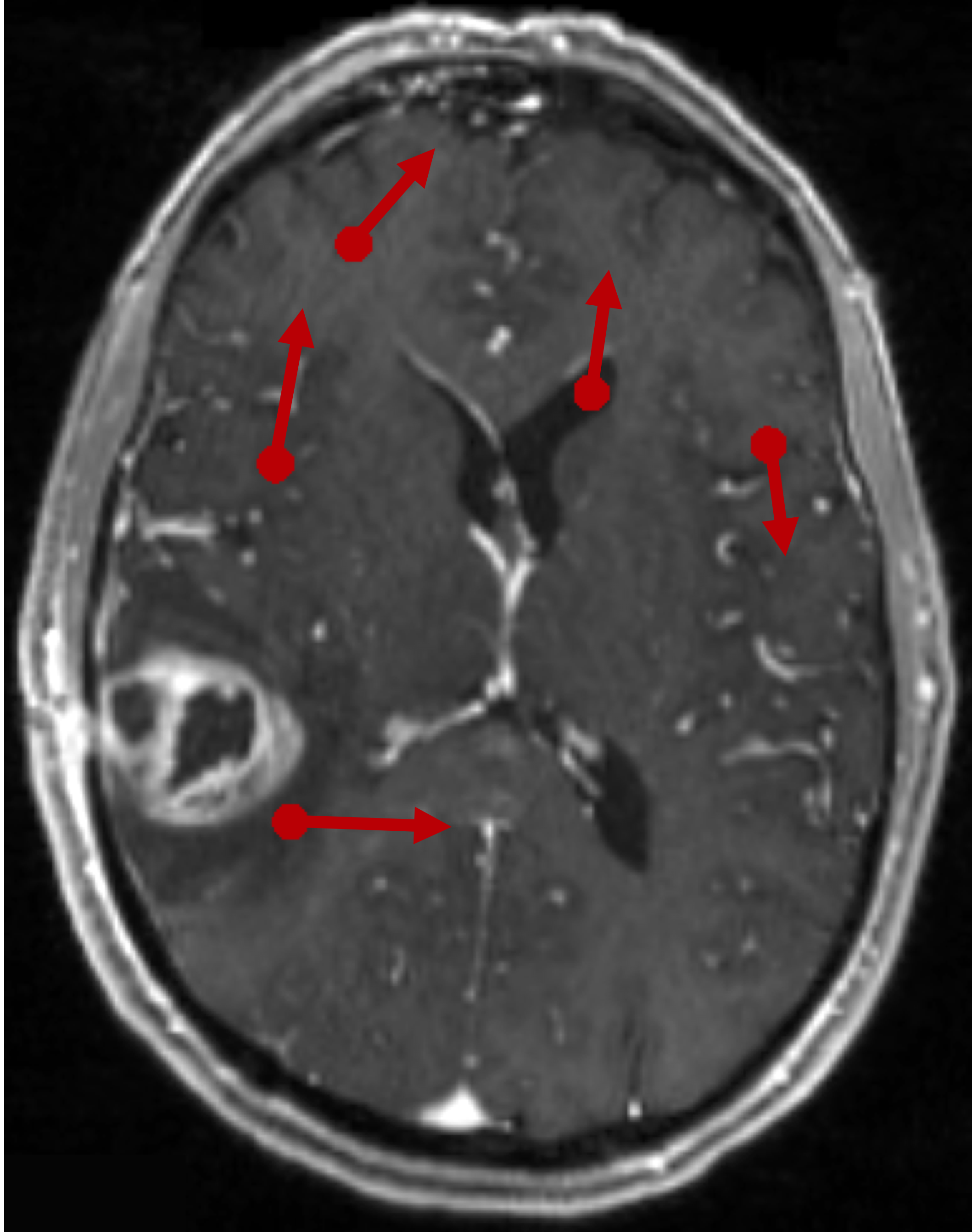
@article{wijethilake2025crossmoda, title = {crossMoDA Challenge: Evolution of Cross-Modality Domain Adaptation Techniques for Vestibular Schwannoma and Cochlea Segmentation from 2021 to 2023}, author = {Wijethilake, Navodini and Dorent, Reuben and Ivory, Marina and Kujawa, Aaron and Cornelissen, Stefan and Langenhuizen, Patrick and Okasha, Mohamed and Oviedova, Anna and Dong, Hexin and Kang, Bogyeong and others}, journal = {arXiv preprint arXiv:2506.12006}, year = {2025}, n_first_authors = {1} } - Optimizing registration uncertainty visualization to support intraoperative decision-making during brain tumor resectionM Geshvadi*, R Dorent, C Galvin, and 8 more authorsInternational Journal of Computer Assisted Radiology and Surgery, 2025
@article{geshvadi2025optimizing, title = {Optimizing registration uncertainty visualization to support intraoperative decision-making during brain tumor resection}, author = {Geshvadi, M and Dorent, R and Galvin, C and Rigolo, L and Haouchine, N and Kapur, T and Pieper, S and Vangel, M and Wells, WM and Golby, AJ and others}, journal = {International Journal of Computer Assisted Radiology and Surgery}, pages = {1--9}, year = {2025}, publisher = {Springer}, } - SegMatch: semi-supervised surgical instrument segmentationMeng Wei*, Charlie Budd, Luis C Garcia-Peraza-Herrera, and 3 more authorsScientific Reports, 2025
Surgical instrument segmentation is recognised as a key enabler in providing advanced surgical assistance and improving computer-assisted interventions. In this work, we propose SegMatch, a semi-supervised learning method to reduce the need for expensive annotation for laparoscopic and robotic surgical images. SegMatch builds on FixMatch, a widespread semi-supervised classification pipeline combining consistency regularization and pseudo-labelling, and adapts it for the purpose of segmentation. In our proposed SegMatch, the unlabelled images are first weakly augmented and fed to the segmentation model to generate pseudo-labels. In parallel, images are fed to a strong augmentation branch and consistency between the branches is used as an unsupervised loss. To increase the relevance of our strong augmentations, we depart from using only handcrafted augmentations and introduce a trainable adversarial augmentation strategy. Our FixMatch adaptation for segmentation tasks further includes carefully considering the equivariance and invariance properties of the augmentation functions we rely on. For binary segmentation tasks, our algorithm was evaluated on the MICCAI Instrument Segmentation Challenge datasets, Robust-MIS 2019 and EndoVis 2017. For multi-class segmentation tasks, we relied on the recent CholecInstanceSeg dataset. Our results show that SegMatch outperforms fully-supervised approaches by incorporating unlabelled data, and surpasses a range of state-of-the-art semi-supervised models across different labelled to unlabelled data ratios.
@article{wei2025segmatch, title = {SegMatch: semi-supervised surgical instrument segmentation}, author = {Wei, Meng and Budd, Charlie and Garcia-Peraza-Herrera, Luis C and Dorent, Reuben and Shi, Miaojing and Vercauteren, Tom}, journal = {Scientific Reports}, volume = {15}, number = {1}, pages = {14042}, year = {2025}, publisher = {Nature Publishing Group UK London}, }
2024
- Learning to Match 2D Keypoints Across Preoperative MR and Intraoperative UltrasoundHassan Rasheed*, Reuben Dorent, Maximilian Fehrentz, and 6 more authorsIn International Workshop on Advances in Simplifying Medical Ultrasound, 2024
We propose in this paper a texture-invariant 2D keypoints descriptor specifically designed for matching preoperative Magnetic Resonance (MR) images with intraoperative Ultrasound (US) images. We introduce a matching-by-synthesis strategy, where intraoperative US images are synthesized from MR images accounting for multiple MR modalities and intraoperative US variability. We build our training set by enforcing keypoints localization over all images then train a patient-specific descriptor network that learns texture-invariant discriminant features in a supervised contrastive manner, leading to robust keypoints descriptors. Our experiments on real cases with ground truth show the effectiveness of the proposed approach, outperforming the state-of-the-art methods and achieving matching precision on average.
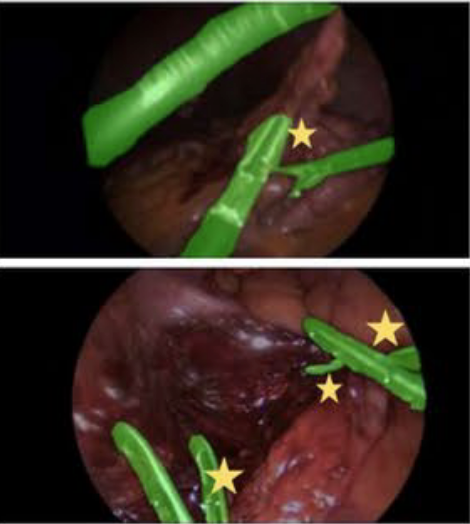
@inproceedings{rasheed2024learning, title = {Learning to Match 2D Keypoints Across Preoperative MR and Intraoperative Ultrasound}, author = {Rasheed, Hassan and Dorent, Reuben and Fehrentz, Maximilian and Kapur, Tina and Wells III, William M and Golby, Alexandra and Frisken, Sarah and Schnabel, Julia A and Haouchine, Nazim}, booktitle = {International Workshop on Advances in Simplifying Medical Ultrasound}, pages = {78--87}, year = {2024}, organization = {Springer}, doi = {10.1007/978-3-031-73647-6_8}, award = {<em><a href="">Best Paper Award</a></em>} } - Patient-specific real-time segmentation in trackerless brain ultrasoundReuben Dorent*, Erickson Torio, Nazim Haouchine, and 5 more authorsIn International Conference on Medical Image Computing and Computer-Assisted Intervention, 2024
Intraoperative ultrasound (iUS) imaging has the potential to improve surgical outcomes in brain surgery. However, its interpretation is challenging, even for expert neurosurgeons. In this work, we designed the first patient-specific framework that performs brain tumor segmentation in trackerless iUS. To disambiguate ultrasound imaging and adapt to the neurosurgeon’s surgical objective, a patient-specific real-time network is trained using synthetic ultrasound data generated by simulating virtual iUS sweep acquisitions in pre-operative MR data. Extensive experiments performed in real ultrasound data demonstrate the effectiveness of the proposed approach, allowing for adapting to the surgeon’s definition of surgical targets and outperforming non-patient-specific models, neurosurgeon experts, and high-end tracking systems. Our code is available at: https://github.com/ReubenDo/MHVAE-Seg.
- Two Projections Suffice for Cerebral Vascular ReconstructionAlexandre Cafaro*, Reuben Dorent, Nazim Haouchine, and 4 more authorsIn International Conference on Medical Image Computing and Computer-Assisted Intervention, 2024
3D reconstruction of cerebral vasculature from 2D biplanar projections could significantly improve diagnosis and treatment planning. We introduce a novel approach to tackle this challenging task by initially backprojecting the two projections, a process that traditionally results in unsatisfactory outcomes due to inherent ambiguities. To overcome this, we employ a U-Net approach trained to resolve these ambiguities, leading to significant improvement in reconstruction quality. The process is further refined using a Maximum A Posteriori strategy with a prior that favors continuity, leading to enhanced 3D reconstructions. We evaluated our approach using a comprehensive dataset comprising segmentations from approximately 700 MR angiography scans, from which we generated paired realistic biplanar DRRs. Upon testing with held-out data, our method achieved an 80% Dice similarity w.r.t the ground truth, superior to existing methods. Our code and dataset are available at https://github.com/Wapity/3DBrainXVascular.
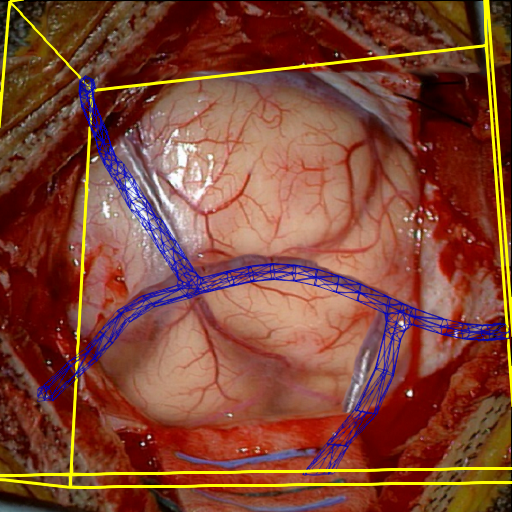
- Intraoperative Registration by Cross-Modal Inverse Neural RenderingMaximilian Fehrentz*, Mohammad Farid Azampour, Reuben Dorent, and 7 more authorsIn International Conference on Medical Image Computing and Computer-Assisted Intervention, 2024
We present in this paper a novel approach for 3D/2D intraoperative registration during neurosurgery via cross-modal inverse neural rendering. Our approach separates implicit neural representation into two components, handling anatomical structure preoperatively and appearance intraoperatively. This disentanglement is achieved by controlling a Neural Radiance Field’s appearance with a multi-style hypernetwork. Once trained, the implicit neural representation serves as a differentiable rendering engine, which can be used to estimate the surgical camera pose by minimizing the dissimilarity between its rendered images and the target intraoperative image. We tested our method on retrospective patients’ data from clinical cases, showing that our method outperforms state-of-the-art while meeting current clinical standards for registration. Code and additional resources can be found at https://maxfehrentz.github.io/style-ngp/.
- Label merge-and-split: A graph-colouring approach for memory-efficient brain parcellationAaron Kujawa*, Reuben Dorent, Sebastien Ourselin, and 1 more authorIn International Conference on Medical Image Computing and Computer-Assisted Intervention, 2024
Whole brain parcellation requires inferring hundreds of segmentation labels in large image volumes and thus presents significant practical challenges for deep learning approaches. We introduce label merge-and-split, a method that first greatly reduces the effective number of labels required for learning-based whole brain parcellation and then recovers original labels. Using a greedy graph colouring algorithm, our method automatically groups and merges multiple spatially separate labels prior to model training and inference. The merged labels may be semantically unrelated. A deep learning model is trained to predict merged labels. At inference time, original labels are restored using atlas-based influence regions. In our experiments, the proposed approach reduces the number of labels by up to 68% while achieving segmentation accuracy comparable to the baseline method without label merging and splitting. Moreover, model training and inference times as well as GPU memory requirements were reduced significantly. The proposed method can be applied to all semantic segmentation tasks with a large number of spatially separate classes within an atlas-based prior.
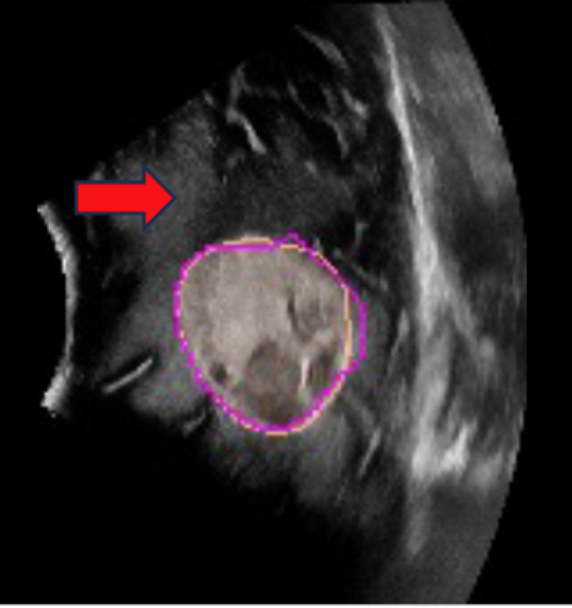
- ReMIND: The Brain Resection Multimodal Imaging DatabaseParikshit Juvekar*, Reuben Dorent*, Fryderyk Kögl*, and 8 more authorsScientific Data, 2024
The standard of care for brain tumors is maximal safe surgical resection. Neuronavigation augments the surgeon’s ability to achieve this but loses validity as surgery progresses due to brain shift. Moreover, gliomas are often indistinguishable from surrounding healthy brain tissue. Intraoperative magnetic resonance imaging (iMRI) and ultrasound (iUS) help visualize the tumor and brain shift. iUS is faster and easier to incorporate into surgical workflows but offers a lower contrast between tumorous and healthy tissues than iMRI. With the success of data-hungry Artificial Intelligence algorithms in medical image analysis, the benefits of sharing well-curated data cannot be overstated. To this end, we provide the largest publicly available MRI and iUS database of surgically treated brain tumors, including gliomas (n = 92), metastases (n = 11), and others (n = 11). This collection contains 369 preoperative MRI series, 320 3D iUS series, 301 iMRI series, and 356 segmentations collected from 114 consecutive patients at a single institution. This database is expected to help brain shift and image analysis research and neurosurgical training in interpreting iUS and iMRI.
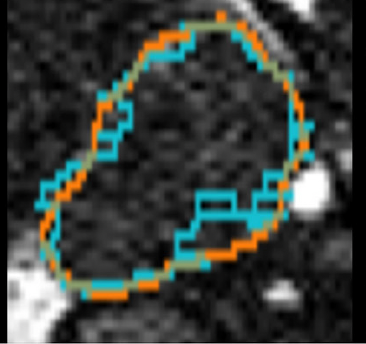
- Deep learning for automatic segmentation of vestibular schwannoma: a retrospective study from multi-center routine MRIAaron Kujawa*, Reuben Dorent, Steve Connor, and 8 more authorsFrontiers in Computational Neuroscience, 2024
Automatic segmentation of vestibular schwannoma (VS) from routine clinical MRI has potential to improve clinical workflow, facilitate treatment decisions, and assist patient management. Previous work demonstrated reliable automatic segmentation performance on datasets of standardized MRI images acquired for stereotactic surgery planning. However, diagnostic clinical datasets are generally more diverse and pose a larger challenge to automatic segmentation algorithms, especially when post-operative images are included. In this work, we show for the first time that automatic segmentation of VS on routine MRI datasets is also possible with high accuracy. We acquired and publicly release a curated multi-center routine clinical (MC-RC) dataset of 160 patients with a single sporadic VS. For each patient up to three longitudinal MRI exams with contrast-enhanced T1-weighted (ceT1w) (n = 124) and T2-weighted (T2w) (n = 363) images were included and the VS manually annotated. Segmentations were produced and verified in an iterative process: (1) initial segmentations by a specialized company; (2) review by one of three trained radiologists; and (3) validation by an expert team. Inter- and intra-observer reliability experiments were performed on a subset of the dataset. A state-of-the-art deep learning framework was used to train segmentation models for VS. Model performance was evaluated on a MC-RC hold-out testing set, another public VS datasets, and a partially public dataset. The generalizability and robustness of the VS deep learning segmentation models increased significantly when trained on the MC-RC dataset. Dice similarity coefficients (DSC) achieved by our model are comparable to those achieved by trained radiologists in the inter-observer experiment. On the MC-RC testing set, median DSCs were 86.2(9.5) for ceT1w, 89.4(7.0) for T2w, and 86.4(8.6) for combined ceT1w+T2w input images. On another public dataset acquired for Gamma Knife stereotactic radiosurgery our model achieved median DSCs of 95.3(2.9), 92.8(3.8), and 95.5(3.3), respectively. In contrast, models trained on the Gamma Knife dataset did not generalize well as illustrated by significant underperformance on the MC-RC routine MRI dataset, highlighting the importance of data variability in the development of robust VS segmentation models. The MC-RC dataset and all trained deep learning models were made available online.
2023
- Why Is the Winner the Best?Matthias Eisenmann*, Annika Reinke*, ..., Reuben Dorent, and 104 more authorsIn Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Jun 2023
International benchmarking competitions have become fundamental for the comparative performance assessment of image analysis methods. However, little attention has been given to investigating what can be learnt from these competitions. Do they really generate scientific progress? What are common and successful participation strategies? What makes a solution superior to a competing method? To address this gap in the literature, we performed a multi-center study with all 80 competitions that were conducted in the scope of IEEE ISBI 2021 and MICCAI 2021. Statistical analyses performed based on comprehensive descriptions of the submitted algorithms linked to their rank as well as the underlying participation strategies revealed common characteristics of winning solutions. These typically include the use of multi-task learning (63%) and/or multi-stage pipelines (61%), and a focus on augmentation (100%), image preprocessing (97%), data curation (79%), and postprocessing (66%). The "typical" lead of a winning team is a computer scientist with a doctoral degree, five years of experience in biomedical image analysis, and four years of experience in deep learning. Two core general development strategies stood out for highly-ranked teams: the reflection of the metrics in the method design and the focus on analyzing and handling failure cases. According to the organizers, 43% of the winning algorithms exceeded the state of the art but only 11% completely solved the respective domain problem. The insights of our study could help researchers (1) improve algorithm development strategies when approaching new problems, and (2) focus on open research questions revealed by this work.
@inproceedings{Eisenmann_2023_CVPR, author = {Eisenmann, Matthias and Reinke, Annika and ... and Dorent, Reuben and Egger, Jan and Ellis, David G. and Engelhardt, Sandy and Ganz, Melanie and Ghatwary, Noha and Girard, Gabriel and Godau, Patrick and Gupta, Anubha and Hansen, Lasse and Harada, Kanako and Heinrich, Mattias P. and Heller, Nicholas and Hering, Alessa and Huaulm\'e, Arnaud and Jannin, Pierre and Kavur, Ali Emre and Kodym, Old\v{r}ich and Kozubek, Michal and Li, Jianning and Li, Hongwei and Ma, Jun and Mart{\'\i}n-Isla, Carlos and Menze, Bjoern and Noble, Alison and Oreiller, Valentin and Padoy, Nicolas and Pati, Sarthak and Payette, Kelly and R\"adsch, Tim and Rafael-Pati\~no, Jonathan and Bawa, Vivek Singh and Speidel, Stefanie and Sudre, Carole H. and van Wijnen, Kimberlin and Wagner, Martin and Wei, Donglai and Yamlahi, Amine and Yap, Moi Hoon and Yuan, Chun and Zenk, Maximilian and Zia, Aneeq and Zimmerer, David and Aydogan, Dogu Baran and Bhattarai, Binod and Bloch, Louise and Br\"ungel, Raphael and Cho, Jihoon and Choi, Chanyeol and Dou, Qi and Ezhov, Ivan and Friedrich, Christoph M. and Fuller, Clifton D. and Gaire, Rebati Raman and Galdran, Adrian and Faura, \'Alvaro Garc{\'\i}a and Grammatikopoulou, Maria and Hong, SeulGi and Jahanifar, Mostafa and Jang, Ikbeom and Kadkhodamohammadi, Abdolrahim and Kang, Inha and Kofler, Florian and Kondo, Satoshi and Kuijf, Hugo and Li, Mingxing and Luu, Minh and Martin\v{c}i\v{c}, Toma\v{z} and Morais, Pedro and Naser, Mohamed A. and Oliveira, Bruno and Owen, David and Pang, Subeen and Park, Jinah and Park, Sung-Hong and Plotka, Szymon and Puybareau, Elodie and Rajpoot, Nasir and Ryu, Kanghyun and Saeed, Numan and Shephard, Adam and Shi, Pengcheng and \v{S}tepec, Dejan and Subedi, Ronast and Tochon, Guillaume and Torres, Helena R. and Urien, Helene and Vila\c{c}a, Jo\~ao L. and Wahid, Kareem A. and Wang, Haojie and Wang, Jiacheng and Wang, Liansheng and Wang, Xiyue and Wiestler, Benedikt and Wodzinski, Marek and Xia, Fangfang and Xie, Juanying and Xiong, Zhiwei and Yang, Sen and Yang, Yanwu and Zhao, Zixuan and Maier-Hein, Klaus and J\"ager, Paul F. and Kopp-Schneider, Annette and Maier-Hein, Lena}, title = {Why Is the Winner the Best?}, booktitle = {Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR)}, month = jun, year = {2023}, pages = {19955-19966}, max_author_limit = {4}, n_first_authors = {1}, } - Generalization Properties of Geometric 3D Deep Learning Models for Medical SegmentationLéo Lebrat*, Rodrigo Santa Cruz*, Reuben Dorent, and 7 more authorsIn 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Apr 2023
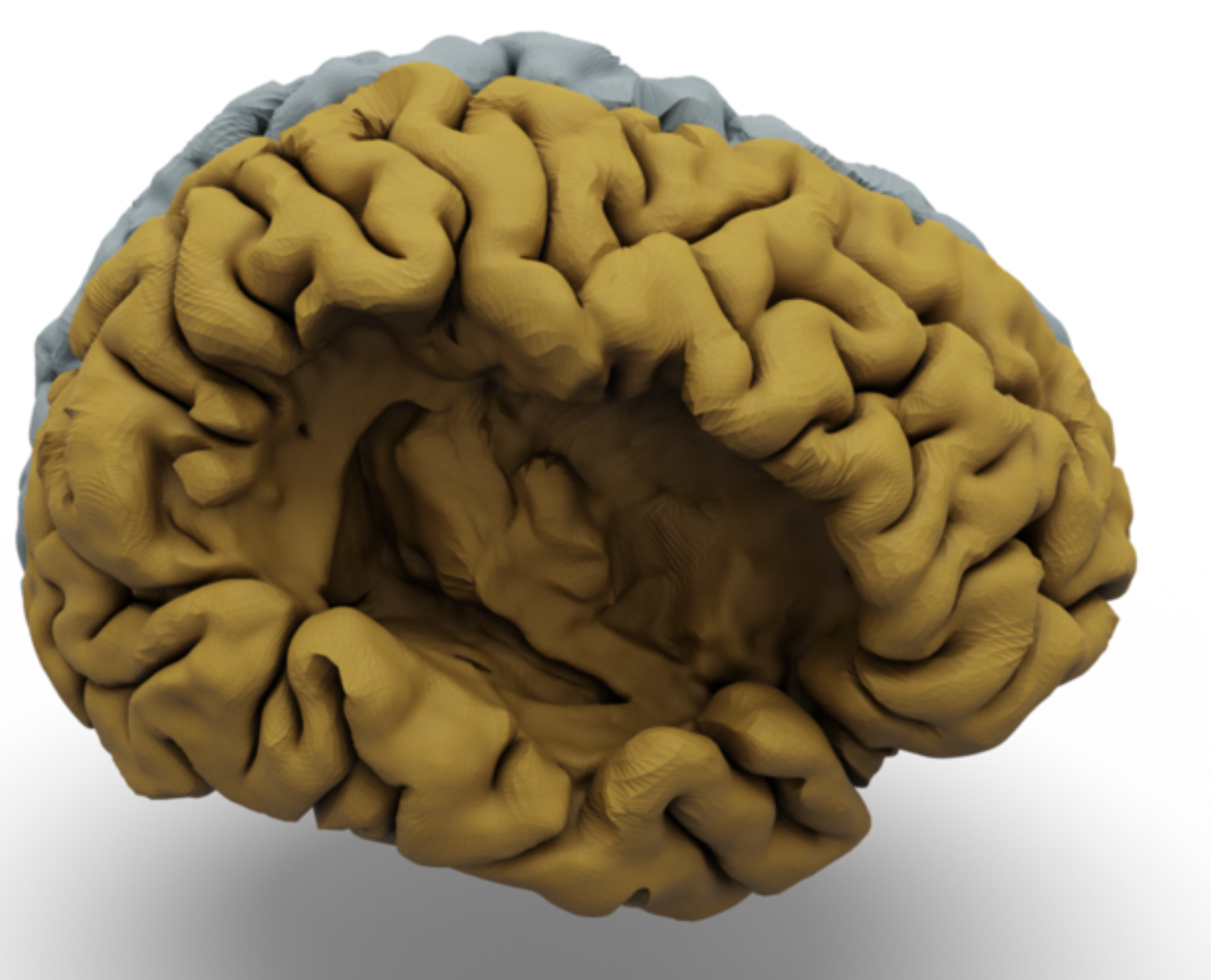
Recent advances in medical Deep Learning (DL) have enabled the significant reduction in time required to extract anatomical segmentations from 3-Dimensional images in an unprecedented manner. Among these methods, supervised segmentation-based approaches using variations of the UNet architecture remain extremely popular. However, these methods remain tied to the input images’ resolution, and their generalisation performance relies heavily on the data distribution over the training dataset. Recently, a new family of approaches based on 3D geometric DL has emerged. These approaches encompass both implicit and explicit surface representation methods and promises to represent a 3D volume using a continuous representation of its surface whilst conserving its topological properties. It has been conjectured that these geometrical methods are more robust to out-of-distribution data and have increased generalisation properties. In this paper, we test these hypotheses for the challenging task of cortical surface reconstruction (CSR) using recently proposed architectures.
@inproceedings{10230549, author = {Lebrat, Léo and Cruz, Rodrigo Santa and Dorent, Reuben and Yaksic, Javier Urriola and Pagnozzi, Alex and Belous, Gregg and Bourgeat, Pierrick and Fripp, Jurgen and Fookes, Clinton and Salvado, Olivier}, booktitle = {2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI)}, title = {Generalization Properties of Geometric 3D Deep Learning Models for Medical Segmentation}, year = {2023}, volume = {}, number = {}, pages = {1-5}, keywords = {}, doi = {10.1109/ISBI53787.2023.10230549}, issn = {1945-8452}, month = apr, n_first_authors = {1}, } - Learning Expected Appearances for Intraoperative Registration During NeurosurgeryNazim Haouchine*, Reuben Dorent, Parikshit Juvekar, and 5 more authorsIn Medical Image Computing and Computer Assisted Intervention – MICCAI 2023, Sep 2023
We present a novel method for intraoperative patient-to-image registration by learning Expected Appearances. Our method uses preoperative imaging to synthesize patient-specific expected views through a surgical microscope for a predicted range of transformations. Our method estimates the camera pose by minimizing the dissimilarity between the intraoperative 2D view through the optical microscope and the synthesized expected texture. In contrast to conventional methods, our approach transfers the processing tasks to the preoperative stage, reducing thereby the impact of low-resolution, distorted, and noisy intraoperative images, that often degrade the registration accuracy. We applied our method in the context of neuronavigation during brain surgery. We evaluated our approach on synthetic data and on retrospective data from 6 clinical cases. Our method outperformed state-of-the-art methods and achieved accuracies that met current clinical standards.
@inproceedings{10.1007/978-3-031-43996-4_22, author = {Haouchine, Nazim and Dorent, Reuben and Juvekar, Parikshit and Torio, Erickson and Wells, William M. and Kapur, Tina and Golby, Alexandra J. and Frisken, Sarah}, editor = {Greenspan, Hayit and Madabhushi, Anant and Mousavi, Parvin and Salcudean, Septimiu and Duncan, James and Syeda-Mahmood, Tanveer and Taylor, Russell}, title = {Learning Expected Appearances for Intraoperative Registration During Neurosurgery}, booktitle = {Medical Image Computing and Computer Assisted Intervention -- MICCAI 2023}, year = {2023}, publisher = {Springer Nature Switzerland}, address = {Cham}, month = sep, pages = {227--237}, isbn = {978-3-031-43996-4}, } - Unified Brain MR-Ultrasound Synthesis Using Multi-modal Hierarchical RepresentationsReuben Dorent*, Nazim Haouchine, Fryderyk Kogl, and 9 more authorsIn Medical Image Computing and Computer Assisted Intervention – MICCAI 2023, Sep 2023
We introduce MHVAE, a deep hierarchical variational auto-encoder (VAE) that synthesizes missing images from various modalities. Extending multi-modal VAEs with a hierarchical latent structure, we introduce a probabilistic formulation for fusing multi-modal images in a common latent representation while having the flexibility to handle incomplete image sets as input. Moreover, adversarial learning is employed to generate sharper images. Extensive experiments are performed on the challenging problem of joint intra-operative ultrasound (iUS) and Magnetic Resonance (MR) synthesis. Our model outperformed multi-modal VAEs, conditional GANs, and the current state-of-the-art unified method (ResViT) for synthesizing missing images, demonstrating the advantage of using a hierarchical latent representation and a principled probabilistic fusion operation. Our code is publicly available (https://github.com/ReubenDo/MHVAE).
@inproceedings{10.1007/978-3-031-43999-5_43, author = {Dorent, Reuben and Haouchine, Nazim and Kogl, Fryderyk and Joutard, Samuel and Juvekar, Parikshit and Torio, Erickson and Golby, Alexandra J. and Ourselin, Sebastien and Frisken, Sarah and Vercauteren, Tom and Kapur, Tina and Wells, William M.}, editor = {Greenspan, Hayit and Madabhushi, Anant and Mousavi, Parvin and Salcudean, Septimiu and Duncan, James and Syeda-Mahmood, Tanveer and Taylor, Russell}, title = {Unified Brain MR-Ultrasound Synthesis Using Multi-modal Hierarchical Representations}, booktitle = {Medical Image Computing and Computer Assisted Intervention -- MICCAI 2023}, year = {2023}, month = sep, publisher = {Springer Nature Switzerland}, address = {Cham}, pages = {448--458}, for_project = {true}, isbn = {978-3-031-43999-5}, award = {<em><a href="">Highlighted Poster</a></em>}, }
2022
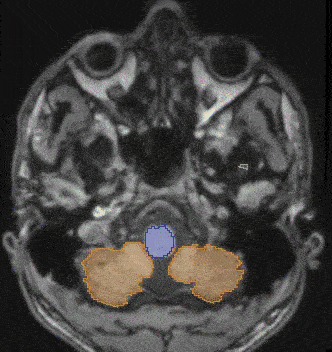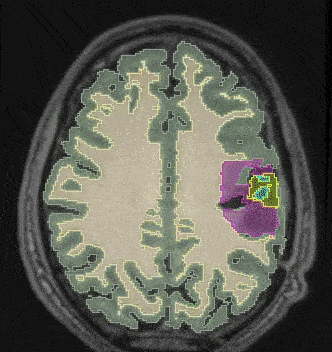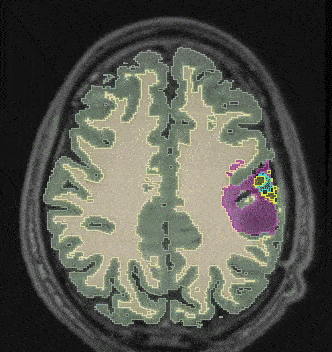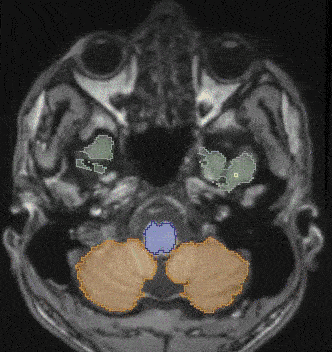
- CrossMoDA 2021 challenge: Benchmark of cross-modality domain adaptation techniques for vestibular schwannoma and cochlea segmentationReuben Dorent*, Aaron Kujawa, Marina Ivory, and 37 more authorsMedical Image Analysis, Oct 2022
Domain Adaptation (DA) has recently been of strong interest in the medical imaging community. While a large variety of DA techniques have been proposed for image segmentation, most of these techniques have been validated either on private datasets or on small publicly available datasets. Moreover, these datasets mostly addressed single-class problems. To tackle these limitations, the Cross-Modality Domain Adaptation (crossMoDA) challenge was organised in conjunction with the 24th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2021). CrossMoDA is the first large and multi-class benchmark for unsupervised cross-modality Domain Adaptation. The goal of the challenge is to segment two key brain structures involved in the follow-up and treatment planning of vestibular schwannoma (VS): the VS and the cochleas. Currently, the diagnosis and surveillance in patients with VS are commonly performed using contrast-enhanced T1 (ceT1) MR imaging. However, there is growing interest in using non-contrast imaging sequences such as high-resolution T2 (hrT2) imaging. For this reason, we established an unsupervised cross-modality segmentation benchmark. The training dataset provides annotated ceT1 scans (N=105) and unpaired non-annotated hrT2 scans (N=105). The aim was to automatically perform unilateral VS and bilateral cochlea segmentation on hrT2 scans as provided in the testing set (N=137). This problem is particularly challenging given the large intensity distribution gap across the modalities and the small volume of the structures. A total of 55 teams from 16 countries submitted predictions to the validation leaderboard. Among them, 16 teams from 9 different countries submitted their algorithm for the evaluation phase. The level of performance reached by the top-performing teams is strikingly high (best median Dice score — VS: 88.4%; Cochleas: 85.7%) and close to full supervision (median Dice score — VS: 92.5%; Cochleas: 87.7%). All top-performing methods made use of an image-to-image translation approach to transform the source-domain images into pseudo-target-domain images. A segmentation network was then trained using these generated images and the manual annotations provided for the source image.
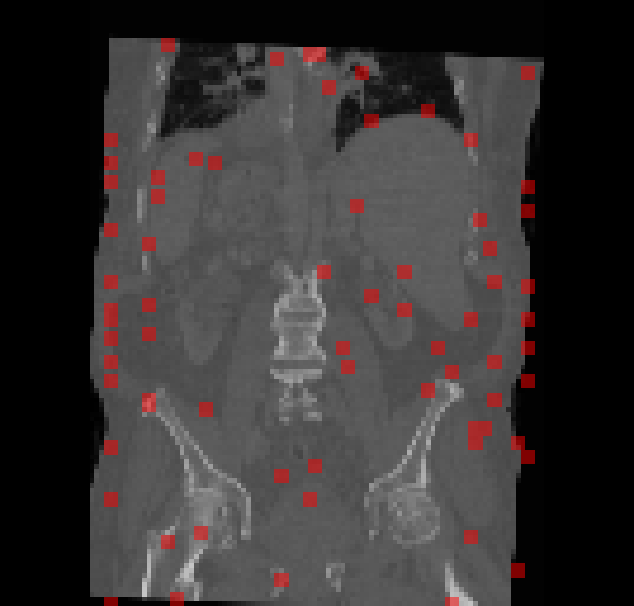
@article{DORENT2023102628, title = {CrossMoDA 2021 challenge: Benchmark of cross-modality domain adaptation techniques for vestibular schwannoma and cochlea segmentation}, journal = {Medical Image Analysis}, volume = {83}, pages = {102628}, year = {2022}, month = oct, issn = {1361-8415}, doi = {10.1016/j.media.2022.102628}, author = {Dorent, Reuben and Kujawa, Aaron and Ivory, Marina and Bakas, Spyridon and Rieke, Nicola and Joutard, Samuel and Glocker, Ben and Cardoso, Jorge and Modat, Marc and Batmanghelich, Kayhan and Belkov, Arseniy and Calisto, Maria Baldeon and Choi, Jae Won and Dawant, Benoit M. and Dong, Hexin and Escalera, Sergio and Fan, Yubo and Hansen, Lasse and Heinrich, Mattias P. and Joshi, Smriti and Kashtanova, Victoriya and Kim, Hyeon Gyu and Kondo, Satoshi and Kruse, Christian N. and Lai-Yuen, Susana K. and Li, Hao and Liu, Han and Ly, Buntheng and Oguz, Ipek and Shin, Hyungseob and Shirokikh, Boris and Su, Zixian and Wang, Guotai and Wu, Jianghao and Xu, Yanwu and Yao, Kai and Zhang, Li and Ourselin, Sébastien and Shapey, Jonathan and Vercauteren, Tom}, keywords = {Domain adaptation, Segmentation, Vestibular schwannoma}, } - Driving Points Prediction for Abdominal Probabilistic RegistrationSamuel Joutard*, Reuben Dorent, Sebastien Ourselin, and 2 more authorsIn Machine Learning in Medical Imaging (MICCAI Workshop), Oct 2022
Inter-patient abdominal registration has various applications, from pharmakinematic studies to anatomy modeling. Yet, it remains a challenging application due to the morphological heterogeneity and variability of the human abdomen. Among the various registration methods proposed for this task, probabilistic displacement registration models estimate displacement distribution for a subset of points by comparing feature vectors of points from the two images. These probabilistic models are informative and robust while allowing large displacements by design. As the displacement distributions are typically estimated on a subset of points (which we refer to as driving points), due to computational requirements, we propose in this work to learn a driving points predictor. Compared to previously proposed methods, the driving points predictor is optimized in an end-to-end fashion to infer driving points tailored for a specific registration pipeline. We evaluate the impact of our contribution on two different datasets corresponding to different modalities. Specifically, we compared the performances of 6 different probabilistic displacement registration models when using a driving points predictor or one of 2 other standard driving points selection methods. The proposed method improved performances in 11 out of 12 experiments.
@inproceedings{10.1007/978-3-031-21014-3_30, author = {Joutard, Samuel and Dorent, Reuben and Ourselin, Sebastien and Vercauteren, Tom and Modat, Marc}, editor = {Lian, Chunfeng and Cao, Xiaohuan and Rekik, Islem and Xu, Xuanang and Cui, Zhiming}, title = {Driving Points Prediction for Abdominal Probabilistic Registration}, booktitle = {Machine Learning in Medical Imaging (MICCAI Workshop)}, year = {2022}, publisher = {Springer Nature Switzerland}, address = {Cham}, pages = {288--297}, month = oct, isbn = {978-3-031-21014-3}, award = {<em><a href="">Best Paper Award</a></em>}, } - Learning joint segmentation of tissues and brain lesions from task-specific hetero-modal domain-shifted datasetsReuben Dorent*, Thomas Booth, Wenqi Li, and 5 more authorsMedical Image Analysis, Jan 2022
Brain tissue segmentation from multimodal MRI is a key building block of many neuroimaging analysis pipelines. Established tissue segmentation approaches have, however, not been developed to cope with large anatomical changes resulting from pathology, such as white matter lesions or tumours, and often fail in these cases. In the meantime, with the advent of deep neural networks (DNNs), segmentation of brain lesions has matured significantly. However, few existing approaches allow for the joint segmentation of normal tissue and brain lesions. Developing a DNN for such a joint task is currently hampered by the fact that annotated datasets typically address only one specific task and rely on task-specific imaging protocols including a task-specific set of imaging modalities. In this work, we propose a novel approach to build a joint tissue and lesion segmentation model from aggregated task-specific hetero-modal domain-shifted and partially-annotated datasets. Starting from a variational formulation of the joint problem, we show how the expected risk can be decomposed and optimised empirically. We exploit an upper bound of the risk to deal with heterogeneous imaging modalities across datasets. To deal with potential domain shift, we integrated and tested three conventional techniques based on data augmentation, adversarial learning and pseudo-healthy generation. For each individual task, our joint approach reaches comparable performance to task-specific and fully-supervised models. The proposed framework is assessed on two different types of brain lesions: White matter lesions and gliomas. In the latter case, lacking a joint ground-truth for quantitative assessment purposes, we propose and use a novel clinically-relevant qualitative assessment methodology.
@article{DORENT2021101862, title = {Learning joint segmentation of tissues and brain lesions from task-specific hetero-modal domain-shifted datasets}, journal = {Medical Image Analysis}, volume = {67}, pages = {101862}, year = {2022}, issn = {1361-8415}, month = jan, doi = {https://doi.org/10.1016/j.media.2020.101862}, url = {https://www.sciencedirect.com/science/article/pii/S1361841520302267}, author = {Dorent, Reuben and Booth, Thomas and Li, Wenqi and Sudre, Carole H. and Kafiabadi, Sina and Cardoso, Jorge and Ourselin, Sebastien and Vercauteren, Tom}, keywords = {Joint learning, Domain adaptation, Multi-Task learning, Multi-Modal}, }
2021
- Inter Extreme Points Geodesics for End-to-End Weakly Supervised Image SegmentationReuben Dorent*, Samuel Joutard, Jonathan Shapey, and 4 more authorsIn Medical Image Computing and Computer Assisted Intervention – MICCAI 2021, Oct 2021
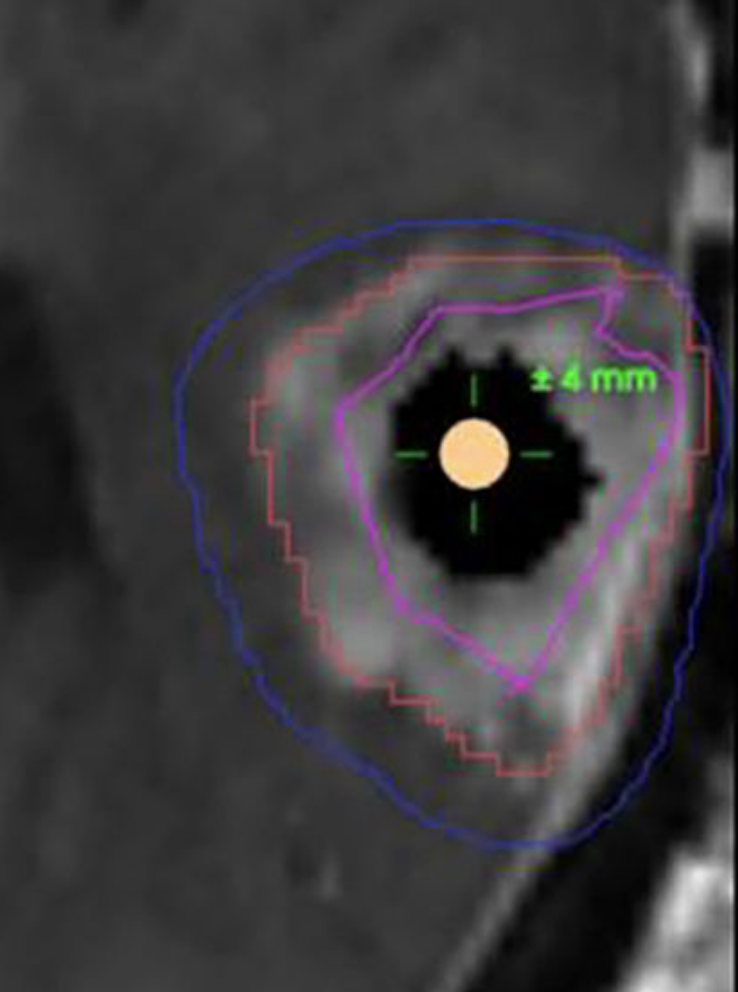
We introduce InExtremIS, a weakly supervised 3D approach to train a deep image segmentation network using particularly weak train-time annotations: only 6 extreme clicks at the boundary of the objects of interest. Our fully-automatic method is trained end-to-end and does not require any test-time annotations. From the extreme points, 3D bounding boxes are extracted around objects of interest. Then, deep geodesics connecting extreme points are generated to increase the amount of “annotated” voxels within the bounding boxes. Finally, a weakly supervised regularised loss derived from a Conditional Random Field formulation is used to encourage prediction consistency over homogeneous regions. Extensive experiments are performed on a large open dataset for Vestibular Schwannoma segmentation. InExtremIS obtained competitive performance, approaching full supervision and outperforming significantly other weakly supervised techniques based on bounding boxes. Moreover, given a fixed annotation time budget, InExtremIS outperformed full supervision. Our code and data are available online.
@inproceedings{10.1007/978-3-030-87196-3_57, author = {Dorent, Reuben and Joutard, Samuel and Shapey, Jonathan and Kujawa, Aaron and Modat, Marc and Ourselin, S{\'e}bastien and Vercauteren, Tom}, editor = {de Bruijne, Marleen and Cattin, Philippe C. and Cotin, St{\'e}phane and Padoy, Nicolas and Speidel, Stefanie and Zheng, Yefeng and Essert, Caroline}, title = {Inter Extreme Points Geodesics for End-to-End Weakly Supervised Image Segmentation}, booktitle = {Medical Image Computing and Computer Assisted Intervention -- MICCAI 2021}, year = {2021}, publisher = {Springer International Publishing}, address = {Cham}, pages = {615--624}, isbn = {978-3-030-87196-3}, award = {<em><a href="">Student Travel Award</a></em>}, month = oct, } - Segmentation of vestibular schwannoma from MRI, an open annotated dataset and baseline algorithmJonathan Shapey*, Aaron Kujawa, Reuben Dorent, and 8 more authorsScientific Data, Oct 2021
Automatic segmentation of vestibular schwannomas (VS) from magnetic resonance imaging (MRI) could significantly improve clinical workflow and assist patient management. We have previously developed a novel artificial intelligence framework based on a 2.5D convolutional neural network achieving excellent results equivalent to those achieved by an independent human annotator. Here, we provide the first publicly-available annotated imaging dataset of VS by releasing the data and annotations used in our prior work. This collection contains a labelled dataset of 484 MR images collected on 242 consecutive patients with a VS undergoing Gamma Knife Stereotactic Radiosurgery at a single institution. Data includes all segmentations and contours used in treatment planning and details of the administered dose. Implementation of our automated segmentation algorithm uses MONAI, a freely-available open-source framework for deep learning in healthcare imaging. These data will facilitate the development and validation of automated segmentation frameworks for VS and may also be used to develop other multi-modal algorithmic models.
@article{shapey2021segmentation, title = {Segmentation of vestibular schwannoma from MRI, an open annotated dataset and baseline algorithm}, author = {Shapey, Jonathan and Kujawa, Aaron and Dorent, Reuben and Wang, Guotai and Dimitriadis, Alexis and Grishchuk, Diana and Paddick, Ian and Kitchen, Neil and Bradford, Robert and Saeed, Shakeel R and others}, journal = {Scientific Data}, volume = {8}, number = {1}, pages = {286}, year = {2021}, publisher = {Nature Publishing Group UK London}, month = oct, } - A self-supervised learning strategy for postoperative brain cavity segmentation simulating resectionsFernando Pérez-Garcı́a*, Reuben Dorent, Michele Rizzi, and 8 more authorsInternational Journal of Computer Assisted Radiology and Surgery, Oct 2021
Accurate segmentation of brain resection cavities (RCs) aids in postoperative analysis and determining follow-up treatment. Convolutional neural networks (CNNs) are the state-of-the-art image segmentation technique, but require large annotated datasets for training. Annotation of 3D medical images is time-consuming, requires highly-trained raters, and may suffer from high inter-rater variability. Self-supervised learning strategies can leverage unlabeled data for training. We developed an algorithm to simulate resections from preoperative magnetic resonance images (MRIs). We performed self-supervised training of a 3D CNN for RC segmentation using our simulation method. We curated EPISURG, a dataset comprising 430 postoperative and 268 preoperative MRIs from 430 refractory epilepsy patients who underwent resective neurosurgery. We fine-tuned our model on three small annotated datasets from different institutions and on the annotated images in EPISURG, comprising 20, 33, 19 and 133 subjects. The model trained on data with simulated resections obtained median (interquartile range) Dice score coefficients (DSCs) of 81.7 (16.4), 82.4 (36.4), 74.9 (24.2) and 80.5 (18.7) for each of the four datasets. After fine-tuning, DSCs were 89.2 (13.3), 84.1 (19.8), 80.2 (20.1) and 85.2 (10.8). For comparison, inter-rater agreement between human annotators from our previous study was 84.0 (9.9). We present a self-supervised learning strategy for 3D CNNs using simulated RCs to accurately segment real RCs on postoperative MRI. Our method generalizes well to data from different institutions, pathologies and modalities. Source code, segmentation models and the EPISURG dataset are available at https://github.com/fepegar/ressegijcars.
@article{perez2021self, title = {A self-supervised learning strategy for postoperative brain cavity segmentation simulating resections}, author = {P{\'e}rez-Garc{\'\i}a, Fernando and Dorent, Reuben and Rizzi, Michele and Cardinale, Francesco and Frazzini, Valerio and Navarro, Vincent and Essert, Caroline and Ollivier, Ir{\`e}ne and Vercauteren, Tom and Sparks, Rachel and others}, journal = {International Journal of Computer Assisted Radiology and Surgery}, volume = {16}, pages = {1653--1661}, year = {2021}, publisher = {Springer}, }
2020
- Scribble-Based Domain Adaptation via Co-segmentationReuben Dorent*, Samuel Joutard, Jonathan Shapey, and 7 more authorsIn Medical Image Computing and Computer Assisted Intervention – MICCAI 2020, Oct 2020
Although deep convolutional networks have reached state-of-the-art performance in many medical image segmentation tasks, they have typically demonstrated poor generalisation capability. To be able to generalise from one domain (e.g. one imaging modality) to another, domain adaptation has to be performed. While supervised methods may lead to good performance, they require to fully annotate additional data which may not be an option in practice. In contrast, unsupervised methods don’t need additional annotations but are usually unstable and hard to train. In this work, we propose a novel weakly-supervised method. Instead of requiring detailed but time-consuming annotations, scribbles on the target domain are used to perform domain adaptation. This paper introduces a new formulation of domain adaptation based on structured learning and co-segmentation. Our method is easy to train, thanks to the introduction of a regularised loss. The framework is validated on Vestibular Schwannoma segmentation (T1 to T2 scans). Our proposed method outperforms unsupervised approaches and achieves comparable performance to a fully-supervised approach.
@inproceedings{10.1007/978-3-030-59710-8_47, author = {Dorent, Reuben and Joutard, Samuel and Shapey, Jonathan and Bisdas, Sotirios and Kitchen, Neil and Bradford, Robert and Saeed, Shakeel and Modat, Marc and Ourselin, S{\'e}bastien and Vercauteren, Tom}, editor = {Martel, Anne L. and Abolmaesumi, Purang and Stoyanov, Danail and Mateus, Diana and Zuluaga, Maria A. and Zhou, S. Kevin and Racoceanu, Daniel and Joskowicz, Leo}, title = {Scribble-Based Domain Adaptation via Co-segmentation}, booktitle = {Medical Image Computing and Computer Assisted Intervention -- MICCAI 2020}, year = {2020}, publisher = {Springer International Publishing}, address = {Cham}, pages = {479--489}, isbn = {978-3-030-59710-8}, month = oct, }
2019
- Hetero-Modal Variational Encoder-Decoder for Joint Modality Completion and SegmentationReuben Dorent*, Samuel Joutard, Marc Modat, and 2 more authorsIn Medical Image Computing and Computer Assisted Intervention – MICCAI 2019, Oct 2019
We propose a new deep learning method for tumour segmentation when dealing with missing imaging modalities. Instead of producing one network for each possible subset of observed modalities or using arithmetic operations to combine feature maps, our hetero-modal variational 3D encoder-decoder independently embeds all observed modalities into a shared latent representation. Missing data and tumour segmentation can be then generated from this embedding. In our scenario, the input is a random subset of modalities. We demonstrate that the optimisation problem can be seen as a mixture sampling. In addition to this, we introduce a new network architecture building upon both the 3D U-Net and the Multi-Modal Variational Auto-Encoder (MVAE). Finally, we evaluate our method on BraTS2018 using subsets of the imaging modalities as input. Our model outperforms the current state-of-the-art method for dealing with missing modalities and achieves similar performance to the subset-specific equivalent networks.
@inproceedings{10.1007/978-3-030-32245-8_9, author = {Dorent, Reuben and Joutard, Samuel and Modat, Marc and Ourselin, S{\'e}bastien and Vercauteren, Tom}, title = {Hetero-Modal Variational Encoder-Decoder for Joint Modality Completion and Segmentation}, booktitle = {Medical Image Computing and Computer Assisted Intervention -- MICCAI 2019}, year = {2019}, publisher = {Springer International Publishing}, address = {Cham}, pages = {74--82}, doi = {10.1007/978-3-030-32245-8_9}, } - Learning joint lesion and tissue segmentation from task-specific hetero-modal datasetsReuben Dorent*, Wenqi Li, Jinendra Ekanayake, and 2 more authorsIn Proceedings of The 2nd International Conference on Medical Imaging with Deep Learning, Jul 2019
Brain tissue segmentation from multimodal MRI is a key building block of many neuroscience analysis pipelines. It could also play an important role in many clinical imaging scenarios. Established tissue segmentation approaches have however not been developed to cope with large anatomical changes resulting from pathology. The effect of the presence of brain lesions, for example, on their performance is thus currently uncontrolled and practically unpredictable. Contrastingly, with the advent of deep neural networks (DNNs), segmentation of brain lesions has matured significantly and is achieving performance levels making it of interest for clinical use. However, few existing approaches allow for jointly segmenting normal tissue and brain lesions. Developing a DNN for such joint task is currently hampered by the fact that annotated datasets typically address only one specific task and rely on a task-specific hetero-modal imaging protocol. In this work, we propose a novel approach to build a joint tissue and lesion segmentation model from task-specific hetero-modal and partially annotated datasets. Starting from a variational formulation of the joint problem, we show how the expected risk can be decomposed and optimised empirically. We exploit an upper-bound of the risk to deal with missing imaging modalities. For each task, our approach reaches comparable performance than task-specific and fully-supervised models.
@inproceedings{pmlr-v102-dorent19a, title = {Learning joint lesion and tissue segmentation from task-specific hetero-modal datasets}, author = {Dorent, Reuben and Li, Wenqi and Ekanayake, Jinendra and Ourselin, Sebastien and Vercauteren, Tom}, booktitle = {Proceedings of The 2nd International Conference on Medical Imaging with Deep Learning}, pages = {164--174}, year = {2019}, editor = {Cardoso, M. Jorge and Feragen, Aasa and Glocker, Ben and Konukoglu, Ender and Oguz, Ipek and Unal, Gozde and Vercauteren, Tom}, volume = {102}, series = {Proceedings of Machine Learning Research}, month = jul, publisher = {PMLR}, url = {https://proceedings.mlr.press/v102/dorent19a.html}, award = {<em><a href="">Long Oral Presentation</a></em>}, } - Permutohedral Attention Module for Efficient Non-local Neural NetworksSamuel Joutard*, Reuben Dorent, Amanda Isaac, and 3 more authorsIn Medical Image Computing and Computer Assisted Intervention – MICCAI 2019, Oct 2019
Medical image processing tasks such as segmentation often require capturing non-local information. As organs, bones, and tissues share common characteristics such as intensity, shape, and texture, the contextual information plays a critical role in correctly labeling them. Segmentation and labeling is now typically done with convolutional neural networks (CNNs) but the context of the CNN is limited by the receptive field which itself is limited by memory requirements and other properties. In this paper, we propose a new attention module, that we call Permutohedral Attention Module (PAM), to efficiently capture non-local characteristics of the image. The proposed method is both memory and computationally efficient. We provide a GPU implementation of this module suitable for 3D medical imaging problems. We demonstrate the efficiency and scalability of our module with the challenging task of vertebrae segmentation and labeling where context plays a crucial role because of the very similar appearance of different vertebrae.
@inproceedings{10.1007/978-3-030-32226-7_44, author = {Joutard, Samuel and Dorent, Reuben and Isaac, Amanda and Ourselin, Sebastien and Vercauteren, Tom and Modat, Marc}, editor = {Shen, Dinggang and Liu, Tianming and Peters, Terry M. and Staib, Lawrence H. and Essert, Caroline and Zhou, Sean and Yap, Pew-Thian and Khan, Ali}, title = {Permutohedral Attention Module for Efficient Non-local Neural Networks}, booktitle = {Medical Image Computing and Computer Assisted Intervention -- MICCAI 2019}, year = {2019}, month = oct, publisher = {Springer International Publishing}, address = {Cham}, pages = {393--401}, doi = {https://doi.org/10.1007/978-3-030-32226-7_44}, isbn = {978-3-030-32226-7}, } - An artificial intelligence framework for automatic segmentation and volumetry of vestibular schwannomas from contrast-enhanced T1-weighted and high-resolution T2-weighted MRIJonathan Shapey*, Guotai Wang*, Reuben Dorent, and 9 more authorsJournal of Neurosurgery JNS, Dec 2019
OBJECTIVE: Automatic segmentation of vestibular schwannomas (VSs) from MRI could significantly improve clinical workflow and assist in patient management. Accurate tumor segmentation and volumetric measurements provide the best indicators to detect subtle VS growth, but current techniques are labor intensive and dedicated software is not readily available within the clinical setting. The authors aim to develop a novel artificial intelligence (AI) framework to be embedded in the clinical routine for automatic delineation and volumetry of VS. METHODS: Imaging data (contrast-enhanced T1-weighted [ceT1] and high-resolution T2-weighted [hrT2] MR images) from all patients meeting the study’s inclusion/exclusion criteria who had a single sporadic VS treated with Gamma Knife stereotactic radiosurgery were used to create a model. The authors developed a novel AI framework based on a 2.5D convolutional neural network (CNN) to exploit the different in-plane and through-plane resolutions encountered in standard clinical imaging protocols. They used a computational attention module to enable the CNN to focus on the small VS target and propose a supervision on the attention map for more accurate segmentation. The manually segmented target tumor volume (also tested for interobserver variability) was used as the ground truth for training and evaluation of the CNN. We quantitatively measured the Dice score, average symmetric surface distance (ASSD), and relative volume error (RVE) of the automatic segmentation results in comparison to manual segmentations to assess the model’s accuracy. RESULTS: Imaging data from all eligible patients (n = 243) were randomly split into 3 nonoverlapping groups for training (n = 177), hyperparameter tuning (n = 20), and testing (n = 46). Dice, ASSD, and RVE scores were measured on the testing set for the respective input data types as follows: ceT1 93.43%, 0.203 mm, 6.96%; hrT2 88.25%, 0.416 mm, 9.77%; combined ceT1/hrT2 93.68%, 0.199 mm, 7.03%. Given a margin of 5% for the Dice score, the automated method was shown to achieve statistically equivalent performance in comparison to an annotator using ceT1 images alone (p = 4e-13) and combined ceT1/hrT2 images (p = 7e-18) as inputs. CONCLUSIONS: The authors developed a robust AI framework for automatically delineating and calculating VS tumor volume and have achieved excellent results, equivalent to those achieved by an independent human annotator. This promising AI technology has the potential to improve the management of patients with VS and potentially other brain tumors.
@article{AnartificialintelligenceframeworkforautomaticsegmentationandvolumetryofvestibularschwannomasfromcontrastenhancedT1weightedandhighresolutionT2weightedMRI, author = {Shapey, Jonathan and Wang, Guotai and Dorent, Reuben and Dimitriadis, Alexis and Li, Wenqi and Paddick, Ian and Kitchen, Neil and Bisdas, Sotirios and Saeed, Shakeel R. and Ourselin, Sebastien and Bradford, Robert and Vercauteren, Tom}, title = {An artificial intelligence framework for automatic segmentation and volumetry of vestibular schwannomas from contrast-enhanced T1-weighted and high-resolution T2-weighted MRI}, journal = {Journal of Neurosurgery JNS}, year = {2019}, month = dec, publisher = {American Association of Neurological Surgeons}, volume = {134}, number = {1}, doi = {https://doi.org/10.3171/2019.9.JNS191949}, pages = {171 - 179}, url = {https://thejns.org/view/journals/j-neurosurg/134/1/article-p171.xml}, n_first_authors = {1}, }